About
Welcome to the official website for the New York meetup of the Learning on Graphs Conference, an annual research conference that covers areas broadly related to machine learning on graphs and geometry.
This event serves as a local branch of the main conference, providing an environment for researchers in this field to convene and foster discussion and social connections.
The main objective of this event is to build an open and diverse community in the greater NYC area with students, professors, and industry researchers interested in machine learning and graphs (e.g., computer science, discrete math, operations research, etc.), geometry (e.g., applied math, physics, neuroscience, biology, etc.), and networks (e.g., network science, social science, etc.).
Explore the event's recorded sessions here.
Learning Meets Geometry, Graphs, and Networks
📅 29th February – 1st March 2024 📍 Jersey City

Important dates:
- Submission deadline: February 25th 2024
- Deadline for registration: February 25th 2024
- Acceptance notification: February 26th 2024
Registration
Registration for this event is open until Nov 14th, 2024. Registration is free but required for attendance.
In addition, participants can optionally submit a poster for presentation during the poster session. Examples of research areas that are within scope for the poster session are described below under Subject Areas.
To register for participation or to submit a poster, please fill out this form.
Subject Areas
The following is a summary of LoG’s focus, which is not exhaustive. If you doubt that your paper fits the venue, feel free to contact logmeetupnyc@gmail.com!
- Expressive graph neural networks
- GNN architectures (e.g., transformers, new positional encodings, etc.)
- Statistical theory on graphs
- Causal inference and causal discovery (e.g., structural causal models, causal graphical models, etc.)
- Geometry processing and optimization
- Robustness and adversarial attacks on graphs
- Combinatorial optimization and graph algorithms
- Graph kernels
- Graph signal processing and spectral methods
- Graph generative models
- Scalable graph learning models and methods
- Graphs for recommender systems
- Knowledge graphs
- Neural manifold
- Self-supervised learning on graphs
- Structured probabilistc inference
- Graph/Geometric ML (e.g., for health applications, security, computer vision, etc.)
- Graph/Geometric ML infrastructures (e.g., datasets, benchmarks, libraries, etc.)
Schedule
The official event schedule is outlined below:
Download a PDF version of the schedule here.
The details for each presentation slot, encompassing presenter names and their respective talk titles, will be made available shortly.
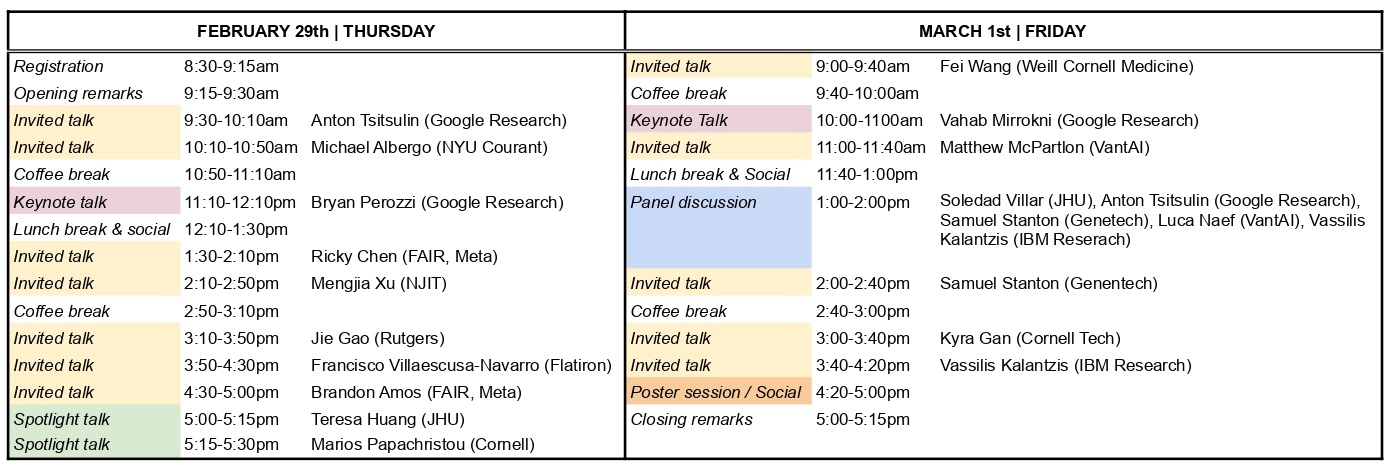
| Time | Slot |
|---|---|
| Thursday 08h30-09h15 | Registration and Reception |
| Thursday 09h15-09h30 | Opening NYC LoG |
| Thursday 09h30-10h10 |  Anton Tsitsulin
Google Research
Anton Tsitsulin
Google Research |
| Thursday 10h10-10h50 |  Michael S Albergo
Courant Institute of Mathematical Sciences
Michael S Albergo
Courant Institute of Mathematical Sciences |
| Thursday 10h50-11h10 | Coffee Break |
| Thursday 11h10-12h10 |  Bryan Perozzi
Google Research
Bryan Perozzi
Google Research |
| Thursday 12h10-13h30 | Lunch Break and Social(lunch not provided) |
| Thursday 13h30-14h10 |  Ricky Tian Qi Chen
FAIR, Meta
Ricky Tian Qi Chen
FAIR, Meta |
| Thursday 14h10-14h50 |  Mengjia Xu
New Jersey Institute of Technology
Mengjia Xu
New Jersey Institute of Technology |
| Thursday 14h50-15h10 | Coffee Break |
| Thursday 15h10-15h50 |  Jie Gao
Rutgers University
Jie Gao
Rutgers University |
| Thursday 15h50-16h30 |  Francisco Villaescusa-Navarro
Flatiron Institute
Francisco Villaescusa-Navarro
Flatiron Institute |
| Thursday 16h30-17h00 |  Brandon Amos
Meta AI (FAIR)
Brandon Amos
Meta AI (FAIR) |
| Thursday 17h00-17h15 | Spotlight Talk |
| Thursday 17h15-17h30 | Spotlight Talk |
| Friday - 09h00-09h40 |  Fei Wang
Weill Cornell Medicine
Fei Wang
Weill Cornell Medicine |
| Friday 9h40-10h00 | Coffee Break |
| Friday - 10h00-11h00 |  Vahab Mirrokni
Google Research
Vahab Mirrokni
Google Research |
| Friday - 11h00-11h40 |  Matthew McPartlon
VantAI
Matthew McPartlon
VantAI |
| Friday - 11h40-13h00 | Lunch Break and Social(lunch not provided) |
| Friday - 13h00-14h00 | Panel Discussion |
| Friday - 14h00-14h40 |  Samuel Stanton
Genetech
Samuel Stanton
Genetech |
| Friday 14h40-15h00 | Coffee Break |
| Friday - 15h00-15h40 |  Kyra Gan
Cornell Tech
Kyra Gan
Cornell Tech |
| Friday - 15h40-16h20 |  Vassilis Kalantzis
IBM Research
Vassilis Kalantzis
IBM Research |
| Friday 16h20-17h00 | Poster Session |
| Friday 17h00-17h15 | Closing Remarks |
Speakers

Thursday 09h30-10h10 Anton Tsitsulin
Advances in Unsupervised Graph Learning
Abstract: We rarely have clear supervision signals in real-world data. Unsupervised learning is even more important to get right for graph-structured data since labeling graphs is a hard task even for competent human labellers. This talk will cover novel ways to do unsupervised graph learning, novel ways of improving structure of graphs, and our approach to benchmarking graph neural networks.
Biography: Anton Tsitsulin is a research scientist at Google Research in NYC. His research interests are scalable, principled methods for analyzing graph data in an unsupervised manner. He received his Ph.D. from the university of Bonn in 2021.

Thursday 10h10-10h50 Michael S Albergo
Measure Transport Perspectives on Sampling, Generative Modeling, and Beyond
Abstract: Both the social and natural world are replete with complex structure that often has a probabilistic interpretation. In the former, we may seek to model, for example, the distribution of natural images or language, for which there are copious amounts of real world data. In the latter, we are given the probabilistic rule describing a physical process, but no procedure for generating samples under it necessary to perform simulation. In this talk, I will discuss a generative modeling paradigm based on maps between probability distributions that is applicable to both of these circumstances. I will describe a means for learning these maps in the context of problems in statistical physics, how to impose symmetries on them to facilitate learning, and how to use the resultant generative models in a statistically unbiased fashion. I will then describe a paradigm that unifies flow-based and diffusion based generative models by recasting generative modeling as a problem of regression. I will demonstrate the efficacy of doing this in computer vision problems and end with some future challenges and applications.
Biography: Michael Albergo is a postdoc at the Courant Institute of Mathematical Sciences. His research interests lie at the intersection of generative modeling and statistical physics, with a focus on designing machine learning methods to advance scientific computing. He received his PhD under the supervision of Kyle Cranmer, Yann LeCun, and Eric Vanden-Eijnden at NYU, his MPhil at the University of Cambridge, and his AB at Harvard University. Starting in August 2024 he will be a Junior Fellow at the Harvard Society of Fellows and an IAIFI Fellow at MIT.

Thursday 11h10-12h10 Bryan Perozzi
Giving a Voice to Your Graph: Data Representations in the LLM Age
Abstract: Graphs are powerful tools for representing complex real-world relationships, essential for tasks like analyzing social networks or identifying financial trends. While large language models (LLMs) have revolutionized natural text reasoning, their application to graphs remains an understudied frontier. To bridge this gap, we need to transform structured graph data into representations LLMs can process. This talk delves into our work on finding the correct graph inductive bias for Graph ML and developing strategies to convert graphs into language-like formats for LLMs. I'll explore our work on “Talking Like a Graph”, and our parameter-efficient method, GraphToken, which learns an encoding function to extend prompts with explicit structured information.
Biography: Bryan Perozzi is a Research Scientist in Google Research’s Algorithms and Optimization group, where he routinely analyzes some of the world’s largest (and perhaps most interesting) graphs. Bryan’s research focuses on developing techniques for learning expressive representations of relational data with neural networks. These scalable algorithms are useful for prediction tasks (classification/regression), pattern discovery, and anomaly detection in large networked data sets. Bryan is an author of 40+ peer-reviewed papers at leading conferences in machine learning and data mining (such as NeurIPS, ICML, ICLR, KDD, and WWW). His doctoral work on learning network representations (DeepWalk) was awarded the prestigious SIGKDD Dissertation Award. Bryan received his Ph.D. in Computer Science from Stony Brook University in 2016, and his M.S. from the Johns Hopkins University in 2011.

Thursday 13h30-14h10 Ricky Tian Qi Chen
Flow Matching on General Geometries
Abstract: In this talk, He will be discussing the problem of defining generative models on manifolds. Designing a general recipe for training probabilistic models that works immediately on a wide selection of manifolds has been a difficult challenge, with some naive approaches ignoring the topology of the manifold, suffering from algorithmic complexities, or resulting in biased training objectives. He will introduce his recent work on Riemannian Flow Matching (RFM), a simple yet powerful framework for training continuous normalizing flows on manifolds, which bypasses many inherent limitations and offers several advantages over previous approaches: it is simulation-free on simple geometries, does not suffer from scaling difficulties, and its objective is a simple regression with closed-form target vector fields. The key ingredient behind RFM is the construction of a relatively simple premetric for defining target vector fields, which generalizes the existing Euclidean case. To extend to general geometries, we propose the use of spectral decompositions to efficiently compute premetrics on the fly. Our method achieves state-of-the-art performance on many real-world non-Euclidean datasets, and we demonstrate tractable training on general geometries, including triangular meshes with highly non-trivial curvature and boundaries.
Biography: Ricky Tian Qi Chen is a Research Scientist at FAIR, Meta based in New York. His research is on building simplified abstractions of the world through the lens of dynamical systems and flows. He generally works on integrating structured transformations into probabilistic modeling, with the goal of improved interpretability, tractable optimization, or extending into novel areas of application.

Thursday 14h10-14h50 Mengjia Xu
Hyperbolic Brain Network Representations for Subjective Cognitive Decline Prediction and Detecting Healthy Brain Aging Trajectories
Abstract: An expansive area of research focuses on discerning patterns of alterations in functional brain networks from the early stages of Alzheimer’s disease, even at the subjective cognitive decline (SCD) stage. Here, we developed a novel hyperbolic MEG brain network embedding framework for transforming complex MEG brain networks into lower-dimensional hyperbolic representations. Using this model, we computed hyperbolic embeddings of the MEG brain networks of two distinct participant groups: individuals with SCD and healthy controls. We demonstrated that these embeddings preserve both local and global geometric information, presenting reduced distortion compared to rival models, even when brain networks are mapped into low-dimensional spaces. In addition, our findings showed that the hyperbolic embeddings encompass unique SCD-related information that improves the discriminatory power above and beyond that of connectivity features alone. Overall, this study presents the first evaluation of hyperbolic embeddings of MEG brain networks, offering novel insights into brain organization, cognitive decline, and potential diagnostic avenues of Alzheimer’s disease.
Biography: Mengjia Xu is currently an Assistant Professor at Department of Data Science, Ying Wu College of Computing, NJIT. She also holds a Research Affiliate position with the MIT NSF Center for Brains, Minds, and Machines (CBMM) at McGovern Institute for Brain Research.

Thursday 15h10-15h50 Jie Gao
Graph Ricci Flow and Applications in Network Analysis and Learning
Abstract: The notion of curvature describes how spaces are bent at each point and Ricci flow deforms the space such that curvature changes in a way analogous to the diffusion of heat. In this talk I will discuss some of our work on discrete Ollivier Ricci curvature defined on graphs. Discrete curvature defined on an edge captures the local connectivity in the neighborhood. In general edges within a densely connected community have positive curvature while edges connecting different communities have negative curvature. By deforming edge weights with respect to curvature one can derive a Ricci flow metric which is robust to edge insertion/deletion. I will present applications of graph Ricci flow in graph analysis and learning, including network alignment, community detection and graph neural networks.
Biography: Jie Gao is a Professor of Computer Science department of Rutgers University. From 2005-2019 she was on faculty of Department of Computer Science, Stony Brook University. Her reserach is in the intersection of Algorithm Design, Computational Geometry and Networking applications such as wireless, mobile, and sensor networks, and more recently social networks, trajectory data/privacy, and scheduling problems in robotics and networking.

Thursday 15h50-16h30 Francisco Villaescusa-Navarro
Learning the laws and composition of the Universe with cosmic graphs
Abstract: Cosmology is a branch of astrophysics dedicated to the study of the laws and constituents of the Universe. To achieve this, cosmologists look at the spatial distribution of galaxies in the Universe with the goal of finding patterns in that distribution that reveal the fundamental physics behind the dynamics of the cosmos. In this talk, I will show how deep learning is revolutionizing the way cosmologists tackle decades-old problems and how graph neural networks can be used in combination with state-of-the-art hydrodynamic simulations to maximize the amount of information that can be extracted from cosmological observations.
Biography: Francisco (Paco) Villaescusa-Navarro is a research scientist at the Flatiron Institute in New York City. He did his PhD at the University of Valencia in Spain. He held postdoctoral positions at the Astronomical Observatory of Trieste, Italy, and the Center for Computational Astrophysics in New York before becoming an associate research scholar at Princeton University, where he holds a visiting research scholar position. Paco is the main architect of the Quijote simulations, the largest suite of cosmological N-body simulations ever run. He is also part of the CAMELS core team that designed and ran the largest set of state-of-the-art hydrodynamic simulations to date. Paco combines the output of numerical simulations with machine learning methods to develop theoretical models to extract the maximum amount of information from cosmological surveys in order to unveil the Universe’s mysteries.

Thursday 16h30-17h00 Brandon Amos
End-to-end learning geometries for graphs, dynamical systems, and regression
Abstract: Every machine learning setting has an underlying geometry where the data is represented and the predictions are performed in. While defaulting the geometry to a Euclidean or known manifold is capable of building powerful models, /learning/ a non-trivial geometry from data is useful for improving the overall performance and estimating unobserved structures. This talk focuses on learning geometries for: 1) *graph embeddings*, where the geometry of the embedding, (e.g., Euclidean, spherical, or hyperbolic) heavily influences the accuracy and distortion of the embedding; 2) *dynamical systems*, where the geometry of the state space can uncover unobserved properties of the underlying systems, e.g., geographic information such as obstacles or terrains; and 3) *regression*, where the geometry of the prediction space influences where the model should be accurate or inaccurate for some downstream task. We will focus on /latent/ geometries in these settings that are not directly observable from the data, i.e., the geometry cannot be estimated as a submanifold of the Euclidean space the data is observed in. Instead, the geometry can be shaped via a downstream signal that propagates through differentiable operations such as the geodesic distance, and log/exp maps on Riemannian manifolds. The talk covers the foundational tools here on making operations differentiable (in general via the envelope and implicit function theorems, and simpler when closed-form operations are available), and demonstrates where the end-to-end learned geometry is effective.
Biography: Brandon Amos is a Research Scientist in Meta AI’s Fundamental AI Research group in NYC. He holds a PhD in Computer Science from Carnegie Mellon University and was supported by the USA National Science Foundation Graduate Research Fellowship (NSF GRFP). Prior to joining Meta, he has worked at Adobe Research, Google DeepMind, and Intel Labs. His research interests are in machine learning and optimization with a recent focus on reinforcement learning, control, optimal transport, and geometry.

Friday - 09h00-09h40 Fei Wang
The Next Generation of AI and Data Science in Computational Health: A Full-Stack Holistic Perspective
Abstract: With the revolution of machine learning technologies in recent years, AI and data science are holding greater promise in understanding diseases and improving quality of care. Computational health is such a research area aiming at developing computational methodologies for deriving insights from various biomedical data. Currently the research in computational health has been mostly siloed, with different communities focusing on analyzing different types of data. However, human health has its own ecosystem with information from all aspects including genome, phenome and exposome. We need to integrate the insights from all of them to have more holistic understandings of diseases. In this talk, I will present the research from my lab health in recent years on building machine learning models for analyzing different types of data involved in different levels of human life science, and the need for transitioning from conventional focused-community based strategy to a holistic full-stack regime for the next-generation health AI/data science research.
Biography: Fei Wang is a Professor in Division of Health Informatics, Department of Population Health Sciences, Weill Cornell Medicine (WCM), Cornell University. He is also the founding director of the WCM institute of AI for Digital Health (AIDH). His major research interest is AI and digital health. He has published more than 350 papers on the top venues of related areas such as ICML, KDD, NIPS, CVPR, AAAI, IJCAI, Nature Medicine, JAMA Internal Medicine, Annals of Internal Medicine, Lancet Digital Health, etc. His papers have received over 29,000 citations so far with an H-index 81. His (or his students’) papers have won 8 best paper (or nomination) awards at top international conferences on data mining and medical informatics. His team won the championship of the AACC PTHrP result prediction challenge in 2022, NIPS/Kaggle Challenge on Classification of Clinically Actionable Genetic Mutations in 2017 and Parkinson's Progression Markers' Initiative data challenge organized by Michael J. Fox Foundation in 2016. Dr. Wang is the recipient of the NSF CAREER Award in 2018, as well as the inaugural research leadership award in IEEE International Conference on Health Informatics (ICHI) 2019. Dr. Wang also received prestigious industry awards such as the Sanofi iDEA Award (2021), Google Faculty Research Award (2020) and Amazon AWS Machine Learning for Research Award (2017, 2019 and 2022). Dr. Wang’s Research has been supported by a diverse set of agencies including NSF, NIH, ONR, PCORI, MJFF, AHA, etc. Dr. Wang is the past chair of the Knowledge Discovery and Data Mining working group in American Medical Informatics Association (AMIA). Dr. Wang is a fellow of AMIA, a fellow of IAHSI, a fellow of ACMI and a distinguished member of ACM.

Friday - 10h00-11h00 Vahab Mirrokni
Keynote Talk
Abstract: TBA
Biography: Vahab Mirrokni is a Google Fellow and VP at Google Research, leading algorithm and optimization research groups at Google. These research teams include: market algorithms, large-scale graph mining, and large-scale optimization. Previously he was a distinguished scientist and senior research director at Google. He received his PhD from MIT in 2005 and his B.Sc. from Sharif University of Technology in 2001. He joined Google Research in 2008, after research positions at Microsoft Research, MIT and Amazon.com. He is the co-winner of best paper awards at KDD, ACM EC, and SODA. His research areas include algorithms, distributed and stochastic optimization, and computational economics. Recently he has been working on various algorithmic problems in machine learning, online optimization and mechanism design, and large-scale graph-based learning .

Friday - 11h00-11h40 Matthew McPartlon
LATENTDOCK: Protein-Protein Docking with Latent Diffusion
Abstract: Interactions between proteins form the basis for many biological processes, and understanding their relationships is an area of active research. Computational approaches offer a way to facilitate this understanding without the burden of expensive and time-consuming experiments. Here, we introduce LATENTDOCK, a generative model for protein-protein docking. Our method leverages a diffusion model operating within a geometrically-structured latent space, derived from an encoder producing roto-translational invariant representations of protein complexes. Critically, it is able to perform flexible docking, capturing both backbone and side-chain conformational changes. Furthermore, our model can condition on binding sites, leading to significant performance gains. Empirical evaluations show the efficacy of our approach over relevant baselines, even outperforming models that do not account for flexibility.
Biography: Matthew McPartlon is a senior research scientist at VantAI where he works on deep learning models for predicting protein-protein and protein-small molecule interactions. Prior to joining VantAI he completed his PhD in computer science at The University of Chicago under Jinbo Xu. His current research is focused on learning invariant representations of 3D geometry and applying this to structure related problems in biology.
Friday - 13h00-14h00 Panel Discussion
Luca Naef(VantAI), Samuel Stanton(Genetech), Anton Tsitsulin(Google Research), Vassilis Kalantzis(IBM Reserach), Soledad Villar(JHU)
Abstract:
Biography:

Friday - 14h00-14h40 Samuel Stanton
Promises and Pitfalls of Geometric Deep Learning for Protein Design
Abstract: Protein design appears to be an ideal application of geometric deep learning, with well-motivated symmetries and inductive biases from physics and structural biology. Indeed, until recently in silico protein design was completely dominated by hand-crafted structural models. Recent successes with sequence models are challenging this paradigm, but many researchers still believe strong geometric inductive biases are essential for generalization in data-scarce regimes. We will review a pragmatic assessment of the strengths and weaknesses of structural and sequence-based methods, and discuss the greatest opportunities and challenges facing structural methods in particular in contemporary protein design problems.
Biography: Samuel Stanton is a Machine Learning Scientist at Genentech, working on ML-driven drug discovery with the Prescient Design team. Prior to joining Genentech, Samuel received his PhD from the NYU Center for Data Science as an NDSEG fellow under the supervision of Dr. Andrew Gordon Wilson. Samuel’s recent work includes core contributions to Genentech’s “lab-in-the-loop” active learning system for molecule lead optimization, as well as basic research on uncertainty quantification and decision-making with machine learning.

Friday - 15h00-15h40 Kyra Gan
A gentle introduction to causal discovery and local causal discovery
Abstract: Causal discovery is crucial for understanding the data-generating mechanism and the causal relationships between variables. Additionally, it has an important application in causal inference in the context of observational studies: it can enable the identification of valid adjustment sets (VAS) for unbiased effect estimation. In this talk, I will introduce graphical models as well as some classical constraint-based causal discovery algorithms. We will see how these methods are notoriously expensive in the nonparametric setting, with exponential time and sample complexity in the worst case. Finally, I will introduce our recent work Local Discovery by Partitioning, a local causal discovery method that is tailored towards VAS identification.
Biography: Kyra Gan is an Assistant Professor of Operations Research and Information Engineering at Cornell Tech. Prior to joining Cornell, She was a postdoctoral fellow in the Department of Harvard Statistics, working with Susan Murphy. She obtained her Ph.D. degree in Operations Research from the Tepper School of Business, Carnegie Mellon University in May 2022. Prior to CMU, She received my BA degrees in Mathematics (with the Ann Kirsten Pokora Prize) and Economics from Smith College in May 2017. Her research interests include adaptive/online algorithm design in personalized treatment (including micro-randomized trials and N-of-1 trials) under constrained settings, robust and efficient inference and causal discovery methods.

Friday - 15h40-16h20 Vassilis Kalantzis
Capturing graph directionality using transformers
Abstract: Most existing graph transformers typically capture distances between graph nodes and do not take edge direction into account. This is a limiting assumption since many graph applications need to exploit sophisticated relationships in graph data, such as time, causality, or generic dependency constraints. We introduce a novel graph transformer architecture that explicitly takes into account the directionality between connected graph nodes. To achieve this, we make use of dual encodings to represent both potential roles, i.e., source or target, of each pair of vertices linked by a directed edge. These encodings are learned by leveraging the latent adjacency information extracted from a directional attention module, localized with $k$-hop neighborhood information. Extensive experiments on synthetic and real graph datasets show that our approach can lead to accuracy gains. We also discuss an exploration in mapping directed graph transformers to quantum computers.
Biography: Vasileios Kalantzis is a Senior Research Scientist at IBM T.J. Watson Research Center, NY. Prior to his current role he was a Herman H. Goldstine Memorial Postdoctoral fellow (2018-2019) and Staff Research Scientist (2019-2023). Vasileios obtained his PhD (2018) and MSc (2016) in Computer Science and Engineering from the University of Minnesota, Twin Cities, and his MSc (2014) and MEng (2011) in Computer Engineering from the University of Patras, Greece. His research interests lie in topics in numerical linear algebra, high-performance computing, graph analytics, and randomized algorithms.
Spotlight Talks
Thursday 17h00-17h15
Rethinking Symmetries in Graph Neural Networks - Speaker: Teresa Huang
Thursday 17h15-17h30
Network Formation and Dynamics Among Multi-LLMs - Speaker: Marios Papachristou
Organizers
Sponsors
The Venue
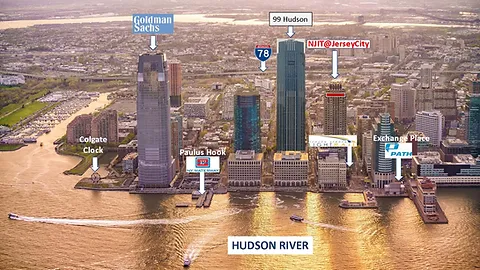
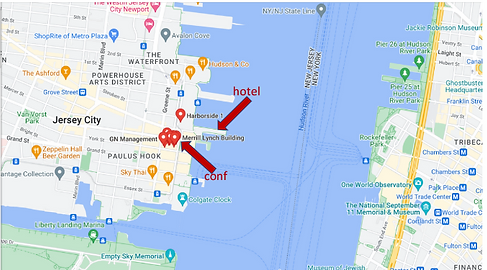
The New York LoG Meet Up will take place in the “NJIT@Jersey City Campus”. The NJIT@JerseyCity is located on the 36th floor of 101 Hudson Street on the Waterfront of Jersey City. The building occupies an entire block bordered by Montgomery Street, Hudson Street, York Street, and Greene Street. “Directions from Google Maps”
Public Transportation
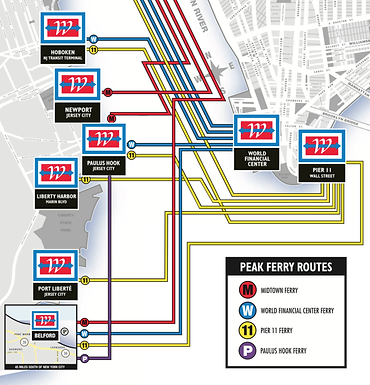
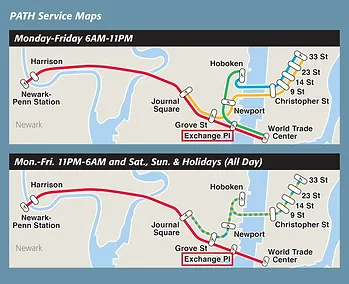
101 Hudson Street is located across the street from Exchange Place station for the PATH and the Hudson-Bergen Light Rail. The PATH at Exchange Place is served by the Newark-World Trade Center and Hoboken-World Trade Center lines. The light rail at Exchange Place is serviced by two routes: the Tonnelle Avenue - West Side Avenue and the Hoboken Terminal - 8th Street service routes. The NY Waterway Paulus Hook Terminal is located at the end of Sussex Street at the Hudson River Waterway three blocks away (10 minutes walking distance) from 101 Hudson Street. There are three ferry routes from NYC: Pier 11/Wall Street., Downtown/Brookfield Place, and Midtown/W. 39th Street.
All building visitors must check in with the security and receive a temporary pass. Walk to the middle elevator bank to access the 36th floor. Exit the elevator and see signs for NJIT@JerseyCity. Turn right to the double glass door entrance to NJIT@JerseyCity (Suite 3610).
Parking and Building Entry
To access the parking garage continue along Hudson Street to York Street; turn right onto York Street and continue towards Greene Street or approach the parking garage by driving along Greene Street and taking a left onto York Street. The entrance to the garage will be on the left.
The entrance to the parking garage is the last "garage door" entrance before the intersection of York and Greene Streets: Google Image of Parking Deck
To enter the parking garage push the button on the ticket dispensing machine; retrieve (and keep!) the ticket to release the arm of the gate to continue driving up the steep ramp to Level 2. Parking is available on Levels 2 - 5. Keep the parking ticket to process payment and to be able to exit the parking deck. Once parked, see signs for the elevators to the lobby.
Exit the garage elevator onto the rear lobby of 101 Hudson Street. Turn right to walk down the corridor towards the front of the building.
All building visitors must check in with the security and receive a temporary pass. Walk to the middle elevator bank to access the 36th floor. Exit the elevator and see signs for NJIT@JerseyCity. Turn right to the double glass door entrance to NJIT@JerseyCity (Suite 3610).